Phage-host interaction
Bacteriophages, short ‘phages’, are viruses preying on bacteria. Since phages represent the most abundant biological entities on this planet, there is virtually no ecosystem where bacteria do not coexist with the viruses infecting them. As model system, we are focusing on phages infecting actinobacteria, representing a phylum of outstanding biotechnological and medical importance (Figure 1).
We aim at a mechanistic understanding of novel components of the bacterial immune system and its potential for the development of innovative biotechnological and medical applications.
The vast majority of antiphage defence systems described until now are mediated by proteins or RNA complexes acting at the cellular level. Recently, we demonstrated that aminoglycosides, a well-known class of antibiotics produced by Streptomyces, are potent inhibitors of phage infection in widely divergent bacterial hosts. We demonstrate that aminoglycosides block an early step of the viral life cycle, prior to genome replication. Our study expands the knowledge of aminoglycoside functions suggesting that aminoglycosides are not only used by their producers as toxic molecules against their bacterial competitors, but could also provide protection against the threat of phage predation at the community level. In ongoing studies, we are aiming at the discovery of novel antiviral molecules produced by bacteria and unravel their function in the context of the bacterial immune system.
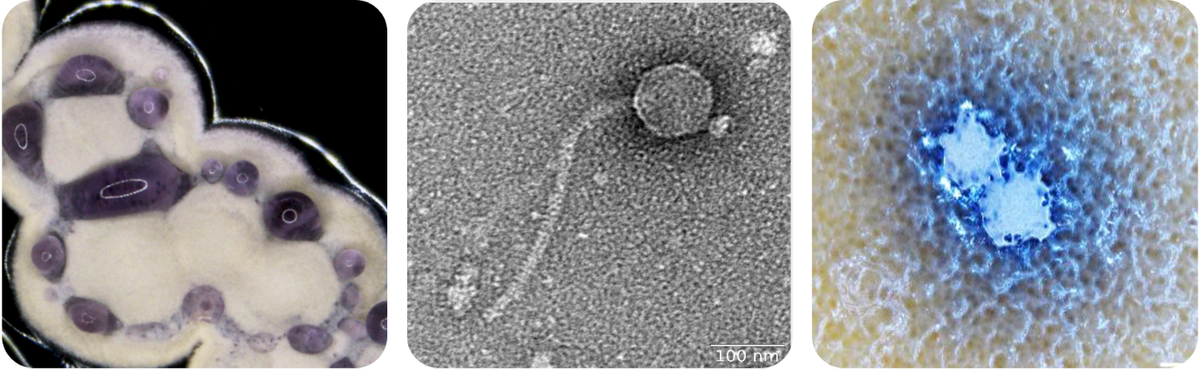
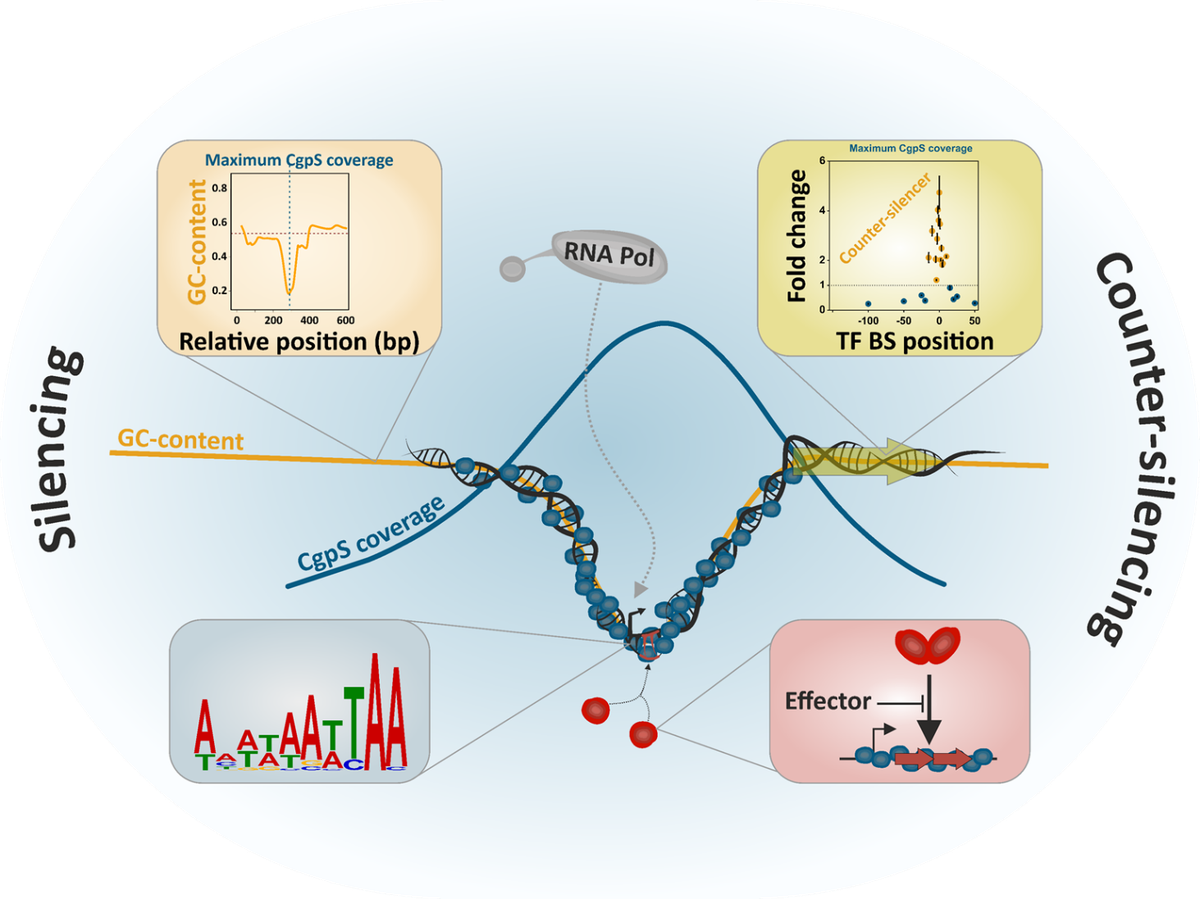
The discrimination between ‘self’ and ‘foreign’ represents a key feature of the bacterial immune system. Besides small molecule-mediated defenses, we study the function and diversity of xenogeneic silencing (XS) proteins. XS proteins, such as the actinobacterial Lsr2-like proteins, can recognize foreign DNA elements by binding to AT-rich DNA regions. These proteins play an important role in the acquisition of novel genetic information and facilitate the evolution of regulatory networks. We recently identified a prophage-encoded Lsr2-like xenogeneic silencing protein (CgpS, Figure 2) playing a key role in the silencing of cryptic prophage islands and for the maintenance of the lysogenic state. Insights are harnessed for the development of novel expression systems or metabolic toggle switches.
Selected publications
Kever L, Hardy A, Luthe T, Hünnefeld M, Gätgens C, Milke L, Wiechert J, Wittmann J, Moraru C, Marienhagen J, Frunzke J (2022) Aminoglycoside antibiotics inhibit phage infection by blocking an early step of the phage infection cycle. bioRxivs preprint, doi.org/10.1101/2021.05.02.442312.
Kever L, Hünnefeld M, Brehm J, Heermann R, Frunzke J. (2021) Identification of Gip as a novel phage-encoded gyrase inhibitor protein of Corynebacterium glutamicum. Mol Microbiol. 116(5):1268-1280. doi: 10.1111/mmi.14813. [FJ1]
Hardy A, Sharma V, Kever L, and Frunzke J (2020) Genome Sequence and Characterization of Five Bacteriophages Infecting Streptomyces Coelicolor and Streptomyces Venezuelae: Alderaan, Coruscant, Dagobah, Endor1 and Endor2. Viruses, doi: https://doi.org/10.3390/v12101065
Wiechert J, Filipchyk A, Hünnefeld M, Gätgens C, Brehm J, Heermann R and Frunzke J, (2020) Deciphering the Rules Underlying Xenogeneic Silencing and Counter-Silencing of Lsr2-like Proteins Using CgpS of Corynebacterium glutamicum as a Model. mBio, doi: 10.1128/mBio.02273-19
Pfeifer E, Michniewski S, Gätgens C, Münch E, Müller F, Polen T, Millard A, Blombach B, and Frunzke J (2019) Generation of a Prophage-Free Variant of the Fast-Growing Bacterium Vibrio natriegens. Appl Environ Microbiol, doi 10.1128/AEM.00853-19
Pfeifer E, Hünnefeld M, Popa O, and Frunzke J (2019) Impact of Xenogeneic Silencing on Phage-Host Interactions. J Mol Biol, doi: 10.1016/j.jmb.2019.02.011
Pfeifer E, Hünnefeld M, Popa O, Polen T, Kohlheyer D, Baumgart M, and Frunzke J (2016) Silencing of cryptic prophages in Corynebacterium glutamicum. Nucleic Acids Res, doi: 10.1093/nar/gkw692sdf
Funding




