Deep Learning for segmentation of 3D-PLI images
Background and Motivation
Three-dimensional Polarized Light Imaging (3D-PLI) technology is used to capture high resolution images of thinly sliced segments of post-mortem brains. These images can then be stacked to reconstruct the brain in three dimensions. However, each image constitutes not only the brain tissue of interest, but also the image background interspersed with the brain tissue.
An accurate 3D reconstruction requires a precise demarcation of the highly irregular border between the brain tissue and the background in each image. Therefore, for each original image, a corresponding binary segmentation mask is required in which each pixel is either marked as brain tissue or background (see Figure 1).


| Figure 1: Large Area Polarimeter (LAP) image of a thin slice of brain (left), and the corresponding segmentation mask (right). |
In terms of anatomical details, the 3D-PLI image dataset consists of two types of images: 1) Images captured with the Large Area Polarimeter (LAP), and 2) Images captured with the Polarizing Microscope (PM). The image shown in Figure 1 is an LAP image of size 2776 by 2080 pixels. The corresponding PM images are typically much larger in size and resolution, e.g., 60,000 by 50,000 pixels.
Our approach
Image segmentation, i.e., the problem of generating a segmentation mask corresponding to an image, constitutes a broad category of problems studied in the field of computer vision. In this project, we are using deep learning based methods in computer vision to develop a solution for the aforementioned segmentation problem for PLI images.
For segmentation of LAP images, we employ the U-Net [1] architecture. Given the size of each LAP image and the number of parameters in the network, it is not possible to simultaneously load the network and the entire input image in GPU memory. Therefore, each image is divided into nine 1024 × 1024 subsections, where each subsection constitutes a single input instance during network training. The network architecture is illustrated in Figure 2.
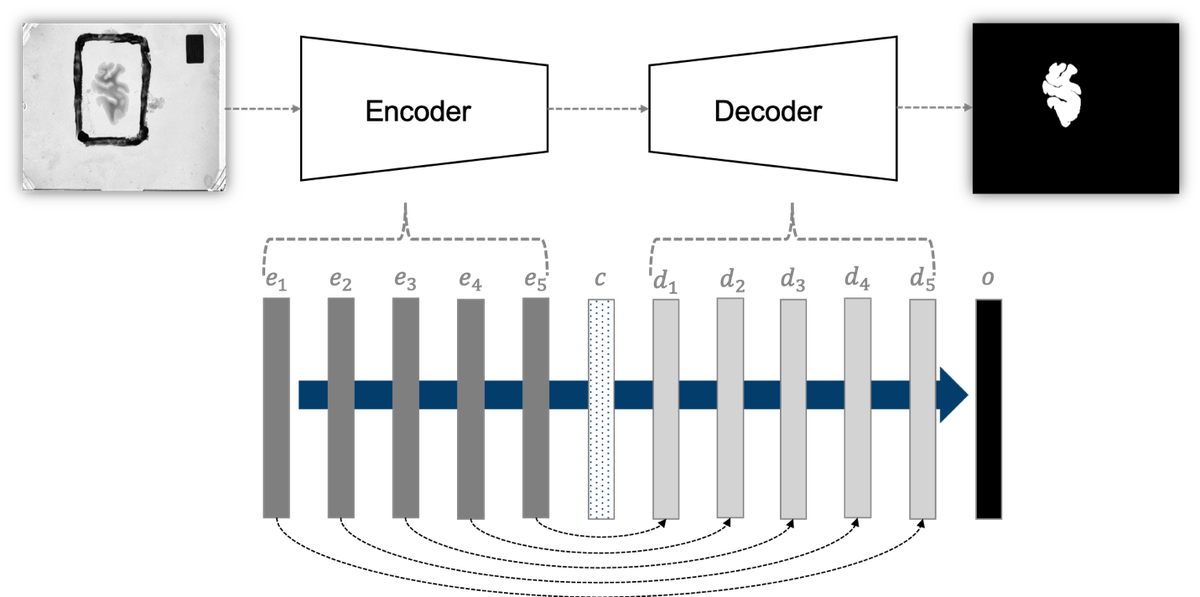
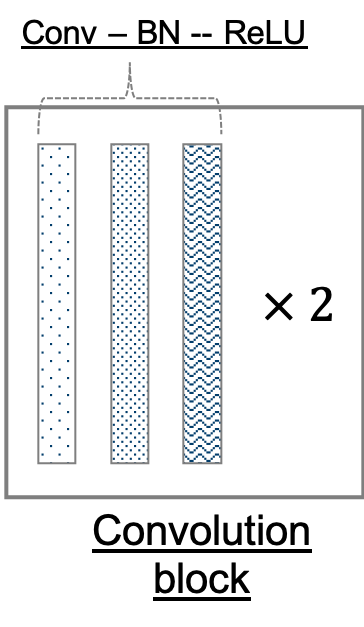

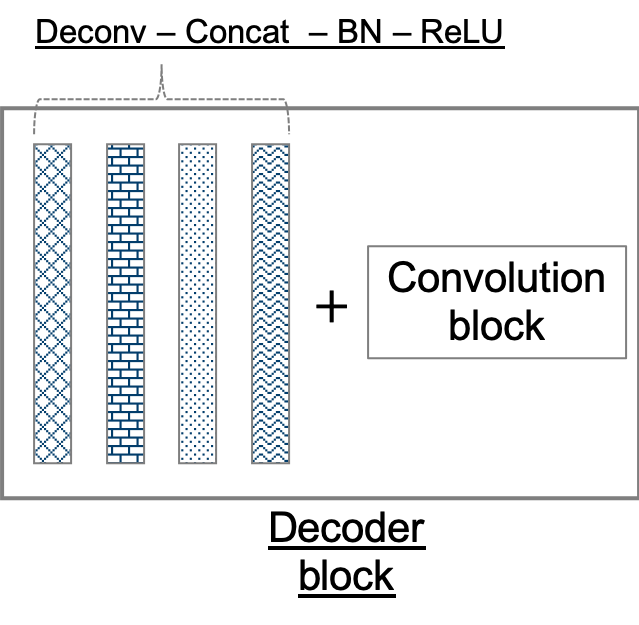
Figure 2: (top) Our adaptation of the U-Net architecture. The network outputs a mask corresponding to the entire input image (an input is a subsection in our application). It comprises of encoder and decoder modules, which further comprise encoder and decoder blocks. Not only are adjacent blocks connected, skip connections are used to directly connect distant encoder and decoder blocks. (bottom left) A convolution block consists of two adjacent sets of convolution, batch normalization, and rectified liner unit layers. (bottom center) An encoder block consists of a convolution block followed by a max pool layer. (bottom right) A decoder block comprises a set of deconvolution, concatenation, batch normalization, and rectified linear unit layers, followed by a convolution block.
The number of properly labeled instances available to us is very small. Therefore, we employ on-the-fly dataset augmentation. In addition to the fairly standard geometric transformations, we employ intensity transformations. A single subsection is then available as input in over thirty different forms.
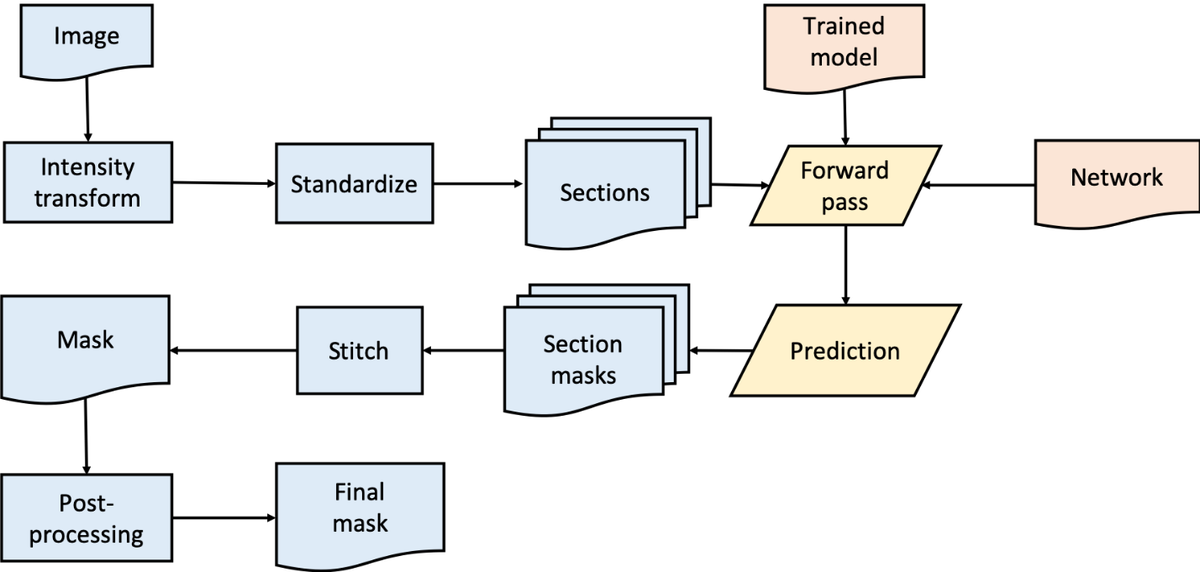
Distributed training on multiple GPUs across multiple nodes on supercomputers is used to speed-up the develop-evaluate-improve cycle. The process of inference for the production environment is presented in Figure 3.
Our work has resulted in models that produce high quality segmentations for the 3D-PLI Large Area Polarimeter images. We are continuing our efforts to further improve the results for deployment in the 3D-PLI workflow. Moreover, we are working on developing solutions for segmentation of Polarizing Microscope (PM) images. The two major challenges there are: 1) the very large image size, and 2) the lack of labeled data.
Our collaboration partners
This project is being conducted in collaboration with the Fiber Architecture group of the INM-1.
Related projects at the Simulation Lab Neuroscience
- Deep Learning for Brain Extraction from MRI scans
- Deep Learning Software for Supercomputers
- hpc4neuro Python library
- Tutorial: Getting started with Deep Learning on Supercomputers
- Using structure tensor analysis to measure PLI image registration quality
- Quality assurance for the 3D-PLI workflow
- Calibration of 3D-PLI measurements
Acknowledgements
The SimLab Neuroscience gratefully acknowledges the computing time granted by the JARA Vergabegremium and provided on the JARA Partition part of the supercomputer JURECA at Forschungszentrum Jülich.
References
- Ronneberger, O., Fischer, P., and Brox, T. “U-Net: Convolutional Networks for Biomedical Image Segmentation”. Medical Image Computing and Computer-Assisted Intervention (MICCAI), Springer, LNCS, Vol.9351: 234—241 (2015).
