VIGAN: Visualization-guided Interactive Graph Analysis
Background and motivation
A vast number of fields in science and engineering, including neuroscience, have benefitted from graph theoretic models. Insights are gained by modeling a system or data as a graph/network, and then performing analyses to identify interesting properties and relations. Such analyses are often based on algorithms borrowed from network science and/or computer science.
In terms of time and effort, this process can be very expensive due to the following reasons:
- A significant amount of programming effort may be required for each project, as often custom scripts need to be written to read and manipulate data, perform analyses, and generate results.
- The larger the amount of code, the more expensive it is to verify code correctness. Test effort alone can be substantial.
In addition, there is the monetary aspect, as some of the most commonly used existing tools for graph analysis in neuroscience rely on software with a proprietary license.
Therefore, a gap exists for a free-to-use tool that minimizes programming and test effort.
Our approach
The Visualization-guided Interactive Graph Analysis (VIGAN) tool is designed to minimize the programming effort required for the exploratory analysis of graph models. We aim to achieve this by providing as many of the standard network analysis algorithms out-of-the-box as possible. Moreover, much of the functionality of VIGAN utilizes the point-and-click method, which alleviates the need to write complex scripts. Also, VIGAN is being developed as free software.
The key features that make VIGAN stand out are:
- User-centered design: A programming-free interface has been a major requirement for VIGAN. Therefore, we have ensured that complex analyses can be carried out without writing any code; merely using point-and-click via the graphical user interface. Nevertheless, for Python-savvy users, all internal data objects are exposed via a Python API that can be accessed via a built-in Python code editor.
- High performance analysis: Graph/Network analysis routines are optimized and parallelized. Therefore, very large datasets can be processed quickly, using massively parallel compute resources available on supercomputers.
- Interactive multi-view visualization: Data can be simultaneously visualized in different formats in multiple-views. Not only does each view support user-interaction for convenient data exploration, all views are optionally interconnected, e.g., clicking a data object in one view can highlight the clicked object in all other connected views. This makes it possible to easily and quickly view data from different perspectives.
- Multi-level analysis: VIGAN provides an interface that allows the user to perform statistical analysis on multiple graphs, and then dive into detailed analysis of individual graphs. This helps find the root cause for effects observed at the level of multi-graph statistical analysis.
- Session recording: VIGAN enables the user to record a video of the application window during a session. Not only does this allow the user to keep a visual record of the steps taken for later reference, the recorded video can also be used as a teaching aid, or sharing the analysis steps with other colleagues.
- Save and resume application state: The user can save the application state at any time and resume work from the same state at a later time; even on a different computer. This makes it possible to continue analysis despite interruptions.
- Extended logging: Provenance tracking is made possible via logging of all user actions per session.
A brief walk-through of interactive exploratory analysis with VIGAN
Consider a scenario where one would like to study functional connectivity using fMRI data. Let us assume that in this case a single vertex in a graph represents aggregated functional activity in a particular region of the brain, while an edge represents the correlation of activity between two regions.
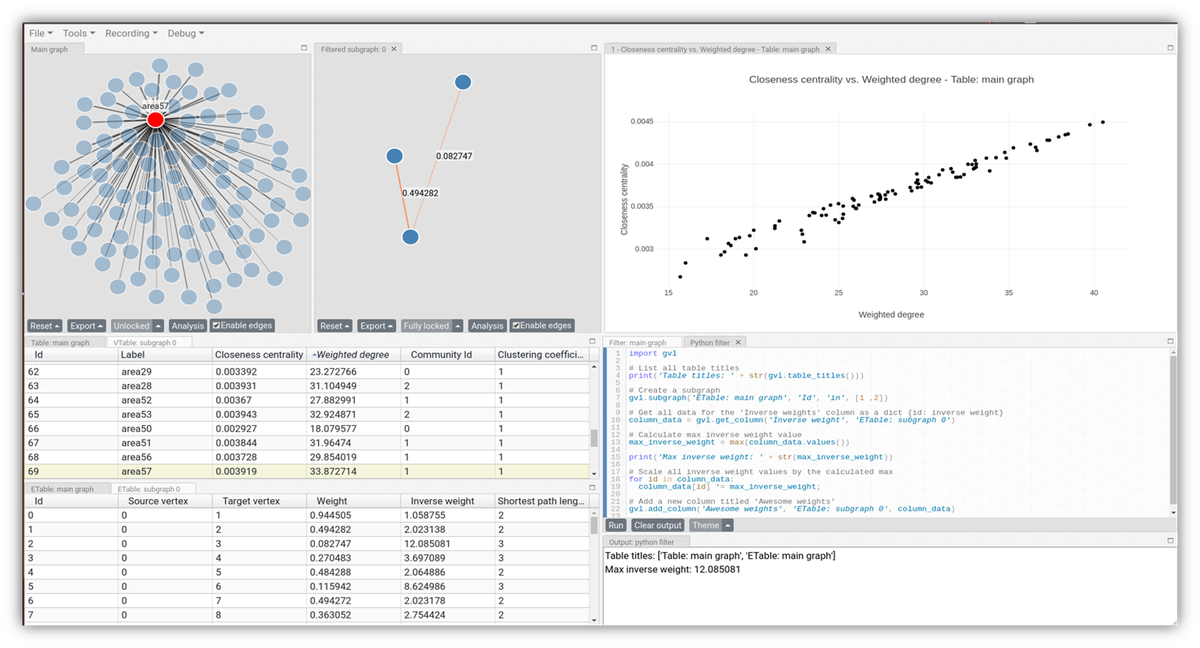
VIGAN allows graph data to be easily loaded via the Graphical User Interface (GUI), along with the option to perform one or more of the several available analyses. Once data has been loaded into the interactive-visualization application, it appears in the graph view (Figure 1, top-left), as well as in the tabular view (Figure 1, bottom-left). For each graph, the tabular view consists of two tables: 1) A table of vertex attributes, and 2) A table of edge attributes. Clicking a vertex in the graph view highlights the corresponding row in the vertex table, and vice versa.
The plot view (Figure 1, top-right) offers the possibility to view data as an interactive chart. E.g., the correlation between closeness centrality and weighted degree in the graph can be viewed as a scatter plot (via a mere couple of mouse clicks). Clicking on a point in the plot highlights the corresponding vertex in the graph view and the corresponding row in the vertex table. This makes it possible to view the data from different perspectives on the same screen.
The bottom-right panel in Figure 1 shows the advanced filter. Here, Python code can be used to retrieve and manipulate VIGAN’s data objects. E.g., a subgraph (Figure 1, top-middle) can be created using a filter criterion to focus the analysis on a specific part of the graph. In addition to the Python filter, a filter based on user interface elements is available.
In the subgraph, shown in the top-middle panel of Figure 1, the highlighted edges represent the shortest path between vertices on either end. Triggering the shortest path analysis is merely a matter of a couple of mouse clicks.
Note: For brevity, only a subset of available features has been presented in the above example.
Current status
We are actively developing VIGAN, and therefore look forward to hearing from interested users. Should you find VIGAN a potentially useful tool for your research, please do let us know.
Our collaboration partners
This application is being developed in collaboration with the Computation in Neural Circuits group of the INM-6.
