Neuroanatomy
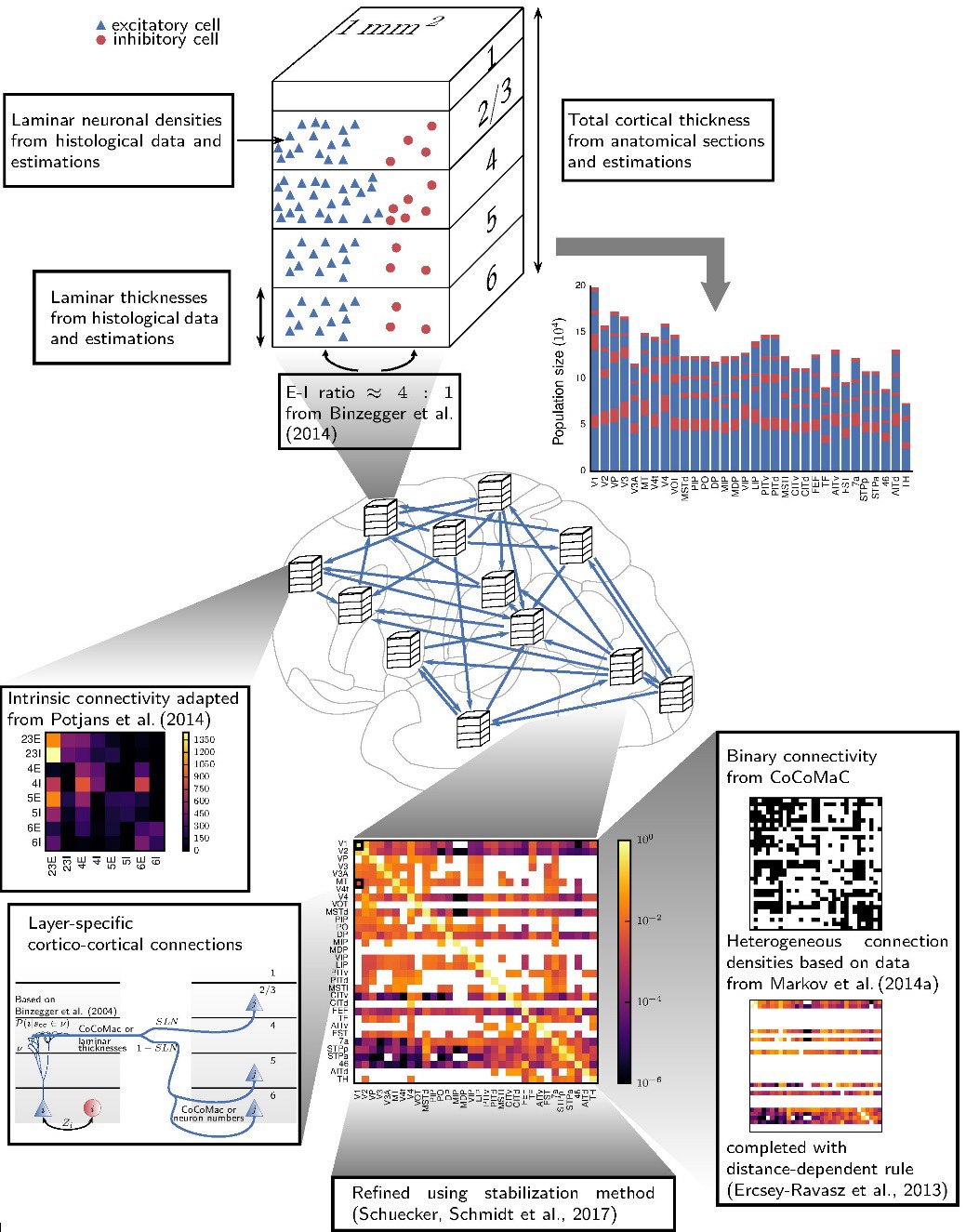
Copyright: Construction principles of a multi-area model of the vision-related areas of macaque cortex (Schmidt et al., 2018)
At the heart of the bottom-up modeling of neuronal networks is the need for a systematic synthesis of anatomical data, in particular cytoarchitectonics (e.g., neuron densities and cortical laminar thicknesses) and the connectivity between neurons. In this part of our work, we bring together such anatomical data from a range of sources, and examine their statistical properties to fill in gaps in the data. Thus, we expose regularities in cortical network structure, both to gain insight into the anatomy, and as an aid toward fully specifying neuronal network models. Furthermore, axonal tracing data for the macaque brain compiled from a large body of experimental studies are made available via CoCoMac, while the Scalable Brain Atlas provides web-based interactive brain atlases of multiple mammalian species.
Publications
- Morales-Gregorio A., van Meegen A., van Albada SJ. (2023) Ubiquitous lognormal distribution of neuron densities across mammalian cerebral cortex. Cereb Cortex 33(16):9439-9449.
DOI: 10.1093/cercor/bhad160 - Timonidis N., Bakker R., Rubio-Teves M., Alonso-Martínez C., Garcia-Amado M., Clascá F., Tiesinga PHE. (2023) Translating single-neuron axonal reconstructions into meso-scale connectivity statistics in the mouse somatosensory thalamus. Front. Neuroinform. 17:1272243.
DOI: 10.3389/fninf.2023.1272243 - Timonidis N., Bakker R., Tiesinga P. (2020) Prediction of a Cell-Class-Specific Mouse Mesoconnectome Using Gene Expression Data. Neuroinformatics 18(4):611-626.
DOI: 10.1007/s12021-020-09471-x - Hilgetag CC., Beul SF., van Albada SJ., Goulas A. (2019) An architectonic type principle integrates macroscopic cortico-cortical connections with intrinsic cortical circuits of the primate brain. Network Neuroscience 3: 905-923.
DOI: 10.1162/netn_a_00100 - Zajzon B., Morales-Gregorio A. (2019) Trans-thalamic Pathways: Strong Candidates for Supporting Communication between Functionally Distinct Cortical Areas. Journal of Neuroscience 39: 7034-7036.
DOI: 10.1523/JNEUROSCI.0656-19.2019 - Schmidt M., Bakker R., Hilgetag CC., Diesmann M., van Albada SJ. (2018) Multi-scale account of the network structure of macaque visual cortex. Brain Structure and Function 223:1409-1435.
DOI: 10.1007/s00429-017-1554-4. - Bezgin G., Solodkin A., Bakker R., Ritter P., McIntosh AR. (2017) Mapping complementary features of cross-species structural connectivity to construct realistic “Virtual Brains”. Human Brain Mapping 38:2080–2093.
DOI: 10.1002/hbm.23506. - Hinne M., Meijers A., Bakker R., Tiesinga PHE., Mørup M., van Gerven MAJ. (2017) The missing link: Predicting connectomes from noisy and partially observed tract tracing data. PLOS Computational Biology 13:e1005374.
DOI: 10.1371/journal.pcbi.1005374. - Bakker R., Tiesinga P., Kötter R. (2015) The Scalable Brain Atlas: Instant Web-Based Access to Public Brain Atlases and Related Content. Neuroinformatics 13:353–366.
DOI: 10.1007/s12021-014-9258-x. - Sergejeva M., Papp EA., Bakker R., Gaudnek MA., Okamura-Oho Y., Boline J., Bjaalie JG., Hess A. (2015) Anatomical landmarks for registration of experimental image data to volumetric rodent brain atlasing templates. Journal of Neuroscience Methods 240:161–169.
DOI: 10.1016/j.jneumeth.2014.11.005. - Tiesinga P., Bakker R., Hill S., Bjaalie JG. (2015) Feeding the human brain model. Current Opinion in Neurobiology 32:107–114.
DOI: 10.1016/j.conb.2015.02.003. - Bezgin G., Rybacki K., van Opstal AJ., Bakker R., Shen K., Vakorin VA., McIntosh AR., Kötter R. (2014) Auditory–prefrontal axonal connectivity in the macaque cortex: Quantitative assessment of processing streams. Brain and Language 135:73–84.
DOI: 10.1016/j.bandl.2014.05.006. - Bakker R., Wachtler T., Diesmann M. (2012) CoCoMac 2.0 and the future of tract-tracing databases. Frontiers in Neuroinformatics 6:30.
DOI: 10.3389/fninf.2012.00030. - Bezgin G., Vakorin VA., van Opstal AJ., McIntosh AR., Bakker R. (2012) Hundreds of brain maps in one atlas: registering coordinate-independent primate neuro-anatomical data to a standard brain. NeuroImage 62:67–76.
DOI: 10.1016/j.neuroimage.2012.04.013.
Book Chapters
- van Albada, SJ., Morales-Gregorio, A., Dickscheid T., Goulas A., Bakker R. Bludau S., Palm G., Hilgetag CC., Diesmann M. (2022). Bringing Anatomical Information into Neuronal Network Models. In: Giugliano, M., Negrello, M., Linaro, D. (eds) Computational Modelling of the Brain. Advances in Experimental Medicine and Biology(), vol 1359. Springer, Cham.
DOI: 10.1007/978-3-030-89439-9_9
Software
CoCoMac: Collation of Connectivity data on the Macaque brain
Scalable Brain Atlas: Web-based interactive brain atlases for macaque, mouse, rat, human, and more
NeuronsReunited neuron database viewer: Viewer for locations and morphologies of long-range projection neurons in mouse
Collaborations
Prof. Timo Dickscheid, INM-1, Forschungszentrum Jülich, Germany
Dr. Sebastian Bludau, INM-1, Forschungszentrum Jülich, Germany
Dr. Christian Schiffer, INM-1, Forschungszentrum Jülich, Germany
Prof. Claus Hilgetag, UKE Hamburg, Germany
Prof. Francisco Clascá, Autonoma University of Madrid, Spain
Prof. Paul Tiesinga, Radboud University, Nijmegen, the Netherlands
Dr. Stefan Mihalas, Allen Institute, USA
Dr. Aitor Morales-Gregorio, Charles University, Czechia
Funding
BMBF (Award ID 2424124) as part of the NSF/NIH/DOE/ANR/BMBF/BSF/NICT/AEI Collaborative Research in Computational Neuroscience Program (2025-2030)
Horizon Europe Programme under the Specific Grant Agreement No. 101147319 (EBRAINS 2.0 Project) (2024-2026)
European Union Horizon 2020 research and innovation program (Grant 945539, Human Brain Project, SGA3) (2020‒2023)
“Cellular, connectional and molecular heterogeneity in a large-scale computational model of the human cerebral cortex“, Deutsche Forschungsgemeinschaft Grant in the Priority Program “Computational Connectomics” (SPP 2041) (2021‒2024)
“Integrating multi-scale connectivity and brain architecture in a large-scale computational model of the human cerebral cortex”, Deutsche Forschungsgemeinschaft Grant in the Priority Program “Computational Connectomics” (SPP 2041) (2018‒2021)
European Union Horizon 2020 research and innovation program (Grant 785907, Human Brain Project, SGA2) (2017‒2020)
