Software
Table of Contents
NEST simulator

The Neural Simulation Tool NEST is a computer program for simulating large heterogeneous networks of point neurons or neurons with a small number of compartments. NEST is best suited for models that focus on the dynamics, size, and structure of neural systems rather than on the detailed morphological and biophysical properties of individual neurons.
Publication: doi:10.4249/scholarpedia.1430
Website: NEST simulator
NESTML

NESTML is a domain-specific language that supports the specification of neuron models in a precise and concise syntax, based on the syntax of Python. Model equations can either be given as a simple string of mathematical notation or as an algorithm written in the built-in procedural language. The equations are analyzed by the associated toolchain, written in Python, to compute an exact solution if possible or use an appropriate numeric solver otherwise.
Publications:
doi:10.3389/fninf.2018.00050
doi:10.5281/zenodo.1319653
doi:10.5281/zenodo.1412345
Website: NESTML
Elephant (Electrophysiology Analysis Toolkit)

Elephant (Electrophysiology Analysis Toolkit) is an open-source, community centered library for the analysis of electrophysiological data in the Python programming language. The focus of Elephant is on generic analysis functions for parallel spike train data and time series recordings, such as the local field potentials (LFP) or intracellular voltages.
Website: Elephant (Electrophisiology Analysis Toolkit)
Neural Microcircuit Simulation and Analysis Toolkit (NMSAT)
Tailor-made python package to build, simulate and analyse neuronal microcircuit models with PyNEST.
Publication: doi:10.5281/zenodo.582645
Website: NMSAT
Image copyright:
GPL, Renato Duarte, Barna Zajzon, & Abigail Morrison. (2017). Neural Microcircuit Simulation and Analysis Toolkit (0.1). Zenodo. https://doi.org/10.5281/zenodo.582645
NetworkUnit

NetworkUnit is a library based on SciUnit to perform model validation testing on the level of the statistics exhibited by the population dynamics of parallel spike and LFP data. Based on capabilities of a given model, it computes common statistical measures on the model and experimental data, and evaluates the level of agreement between the two based on comparing the measures.
Publications:
doi:10.3389/fninf.2018.00090
doi:10.1016/j.biosystems.2022.104813
Website: NetworkUnit
odMLtables

odMLtables is a tool to support working with metadata collections for electrophysiological data. It provides a set of library functions as well as a graphical user interface that offers to switch between hierarchical and flat (tablular) representations of their metadata collection, and provides corresponding functions that assist in working with odML and spreadsheet files.
Publication: doi:10.3389/fninf.2019.00062
Website: odMLtables
Meanfield Toolbox
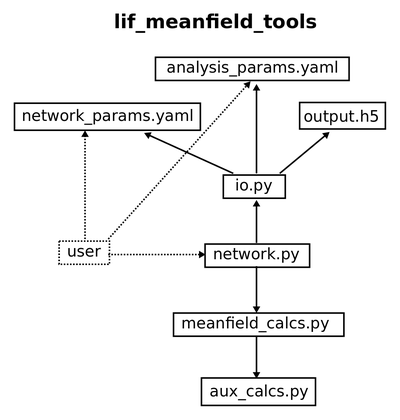
Using this package, you can easily calculate quantities like firing rates, power spectra, and many more, which give you a deeper and more intuitive understanding of what your network does. If your network is not behaving the way you want it to, these tools might help you to figure out, or figure out, or even tell you, what you need to change in order to achieve the desired behaviour. It is easy to store (and in the future, to plot) results and reuse them for further analyses.
Publications:
arXiv:1410.8799
doi:10.1371/journal.pcbi.1005179
doi:10.1103/PhysRevE.92.052119
doi:10.1371/journal.pcbi.1005132
Website: Meanfield Toolbox
Image copyright: Layer, Moritz, Senk, Johanna, Essink, Simon, Korvasová, Karolína, van Meegen, Alexander, Bos, Hannah, … Helias, Moritz. (2020, February 10). LIF Meanfield Tools (Version v0.2). Zenodo. http://doi.org/10.5281/zenodo.3661413; Courtesy- Moritz Helias
beNNch
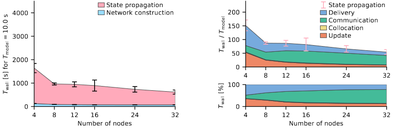
beNNch is a software framework implementing a unified, modular workflow for configuring, executing, and analyzing performance benchmarks of neuronal network simulations on high-performance computing systems.
Publication: https://doi.org/10.3389/fninf.2022.837549
Website: beNNch
Image Copyright: Figure 5C of Albers J., Pronold J., Kurth AC., Vennemo SB., Haghighi Mood K., Patronis A., Terhorst D., Jordan J., Kunkel S., Tetzlaff T., Diesmann M., Senk J. (2022) A Modular Workflow for Performance Benchmarking of Neuronal Network Simulations. Frontiers in Neuroinformatics 16:837549.
Runtime Network Construction on GPU Devices for Large-Scale Spiking Models Archive
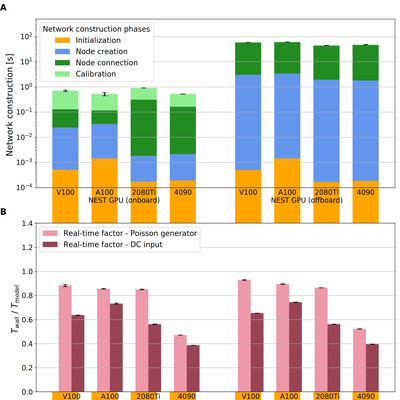
Simulation speed matters for neuroscientific research: this includes not only how fast the simulated model time of a large-scale spiking neuronal network progresses, but also how long it takes beforehand to instantiate the network model in computer memory. On the hardware side, acceleration via highly parallel GPUs is being increasingly utilized. On the software side, code generation approaches ensure highly optimized code, yet on the cost of repeated code regeneration and recompilation after modifications to the network model. Aiming for a greater flexibility with respect to iterative model changes, we here propose a new method for creating network connections interactively, dynamically, and directly in GPU memory through a set of commonly used high-level connection rules. We validate the simulation performance with both consumer and data center GPUs on two neuroscientifically relevant models: a cortical microcircuit of about 77, 000 leaky-integrate-and-fire neuron models and 300 million static synapses, and a balanced random network of excitatory and inhibitory Izhikevich neuron models interconnected with synapses using spike-timing-dependent plasticity. With the proposed ad hoc network construction, both instantiation and simulation times are comparable or even shorter than those obtained with other state-of-the-art simulation technologies, while meeting the flexibility demands of explorative network modeling. Simulation speed matters for neuroscientific research: this includes not only how fast the simulated model time of a large-scale spiking neuronal network progresses, but also how long it takes beforehand to instantiate the network model in computer memory. On the hardware side, acceleration via highly parallel GPUs is being increasingly utilized. On the software side, code generation approaches ensure highly optimized code, yet on the cost of repeated code regeneration and recompilation after modifications to the network model. Aiming for a greater flexibility with respect to iterative model changes, we here propose a new method for creating network connections interactively, dynamically, and directly in GPU memory through a set of commonly used high-level connection rules. We validate the simulation performance with both consumer and data center GPUs on two neuroscientifically relevant models: a cortical microcircuit of about 77, 000 leaky-integrate-and-fire neuron models and 300 million static synapses, and a balanced random network of excitatory and inhibitory Izhikevich neuron models interconnected with synapses using spike-timing-dependent plasticity. With the proposed ad hoc network construction, both instantiation and simulation times are comparable or even shorter than those obtained with other state-of-the-art simulation technologies, while meeting the flexibility demands of explorative network modeling.
Publication: doi:10.3390/app13179598
Website: Runtime Network Construction on GPU Devices for Large-Scale Spiking Models Archive
Image Copyright: Bruno Golosio, Jose Villamar, Gianmarco Tiddia, Elena Pastorelli, Jonas Stapmanns, Viviana Fanti, Pier Stanislao Paolucci, Abigail Morrison, & Johanna Senk. (2023). Runtime Network Construction on GPU Devices for Large-Scale Spiking Models Archive (1.0). Zenodo. https://doi.org/10.5281/zenodo.8304491
